Quick Start#
Import#
To import mapof-roommates python package:
import mapof.roommates as mapof
General Tasks#
Generate SR Instance from Statistical Culture#
In this section, you will learn how to generate stable roommates instances from different statistical cultures. We will start by defining what we mean by a stable roommates instances. Formally, a stable roommates (SR) instance consists of a set of agents, with each agent having a preference order over all other agents.
In practice, we represent an SR instance by RoommatesInstance object, which among other fields, contains:
instance.num_agents # number of agents
instance.votes # preference orders
By votes, we refer to a two-dimensional array list[list[int]], where each row represents a single agent (i.e., vote).
E.g., votes = [[1,2,3],[2,0,3],[3,1,0],[0,2,1] refers to an instance with four following votes:
1 ≻ 2 ≻ 3
2 ≻ 0 ≻ 3
3 ≻ 1 ≻ 0
0 ≻ 2 ≻ 1
instance = mapof.generate_roommates_instance(culture_id='impartial', num_agents=10)
The list of all implemented cultures can be found in List of Cultures.
If you want to add your own culture, you can do this by using the add_culture() function.
experiment.add_culture("my_name", my_func)
The function takes two arguments: The first one is the name of the new culture,
and the second one is the function that generates the votes.
The function that generates the votes can take any number of arguments,
but the first one should be num_agents parameter.
Moreover, the function should return votes as a list of list.
Technically the signature is as follows:
my_func(num_agents: int, **kwargs) -> list[list[int]]:
Compute Distance between Two SR Instances#
To compute a distance, use the compute_distance function, which takes two instances and a distance_id as input.
distances, mapping = mapof.compute_distance(
instance_1,
instance_2,
distance_id='l1-mutual_attraction')
This function returns a tuple containing the distance and the mapping that witnesses this distance. If a given distance does not use a mapping, it returns None instead.
instance_1 = mapof.generate_roommates_instance(culture_id='impartial', num_agents=6)
instance_2 = mapof.generate_roommates_instance(culture_id='impartial', num_agents=6)
distance, mapping = mapof.compute_distance(
instance_1,
instance_2,
distance_id='l1-mutual_attraction')
A list of all implemented distances is available in List of Distances.
Experiments#
Generate SR Instances as Part of Experiment#
In this section, we introduce an abstract object called Experiment, which helps us keep things clear. Finally, we generate instances using the Experiment object.
An Experiment is an abstract object, which, for now, can be seen as a black box in which all the computation takes place. At first, it might be confusing, but in the long run, it simplifies things.
Before carrying out any other operations we need to create an empty Experiment.
For this, we use the function prepare_online_ordinal_experiment(),
which returns an empty Experiment. So, in order to prepare an empty Experiment, type:
experiment = mapof.prepare_online_roommates_experiment()
To give you a hint of what the Experiment is, we present some of its fields and methods:
experiment.instances
experiment.distances
experiment.coordinates
experiment.features
experiment.add_instance()
experiment.add_family()
experiment.compute_distances()
experiment.embed_2d()
experiment.compute_feature()
experiment.print_map_2d()
Now, we will focus on the add_instance() method. In order to generate an instance,
it suffices to run the add_instance() method, and specify the culture_id.
For example, if we want to generate an instance from impartial culture, we type:
experiment.add_instance(culture_id='impartial')
All instances added to the experiment are stored in an experiment.instances dictionary, where the key is the instance_id, and the value is the Instance object. If you want to specify your own instance_id, you can do so using the instance_id argument, for example:
experiment.add_instance(culture_id='impartial', instance_id='IC')
By default, the generated instance will have 20 agents. However, if you want to generate an instance with a different number of agents, use the num_agents argument:
experiment.add_instance(culture_id='impartial', num_agents=20)
If you want to change the default value not for a single SR instance, but for all instances generated in the future, type:
experiment.set_default_num_agents(20)
experiment = mapof.prepare_online_roommates_experiment()
experiment.add_instance(culture_id='impartial', num_agents=16)
Generate Family of SR Instances#
If you would like to add many instances from the same culture, instead of adding them one by one, you can add them as one family of instances.
experiment.add_family(culture_id='impartial', size=10)
The main difference between add_instance and add_family is the fact that the latter function has an additional argument called size,
which specifies how many instances from a given distribution will be created.
Moreover, note that instead of impartial culture, we want to generate Normalized Mallows instances, which are parameterized by norm-ϕ To generate a single Normalized Mallows instance with norm-ϕ = 0.5, we should type:
experiment.add_instance(culture_id='norm-mallows', normphi=0.5)
experiment = mapof.prepare_online_ordinal_experiment()
experiment.add_family(culture_id='norm-mallows', size=10, normphi=0.5)
Create Map of SR Instances#
Creating a map of SR instances is an ultimate tool of this package. We divide the procedure into four major steps, which we describe in detail one by one, with the exception of the first step which was described before. The steps are the following:
Generate instances
Compute distances
Embed in 2D
Print the map
Compute Distances
In order to compute distances between instances, use the following function:
experiment.compute_distances(distance_id='l1-mutual_attraction')
The distances are stored in the distances field, which is a dictionary of dictionaries.
If you want to access the distances, just type:
experiment.distances
Example
Let us assume that we have three instances generated from impartial culture with the following ids: ic_0, ic_1, ic_2.
Then, the distances (dictionary of dictionaries) look as follows:
{'ic_0': {'ic_1': 2.3, 'ic_2': 1.7},
'ic_1': {'ic_0': 2.3, 'ic_2': 1.9},
'ic_2': {'ic_0': 1.7, 'ic_1': 1.9}}
Embedding
In order to embed the instances into 2D Euclidean space, run:
experiment.embed_2d(embedding_id='kk')
The coordinates are stored in the coordinates field, which is a dictionary of lists. If you want to access the coordinates, just type:
experiment.coordinates
More information about different embedding algorithms is available in TBU.
Example
Let us assume that we have four instances generated from Normalized Mallows culture with the following ids: mal_0, mal_1, mal_2, mal_3. Then, the coordinates (dictionary of lists) look as follows:
{'mal_1': [0.2, 0.8],
'mal_2': [0.4, 0.4],
'mal_3': [0.3, 0.1],
'mal_4': [0.9, 0.7]}
Printing
In order to print the map, run:
experiment.print_map_2d()
Example 1.#
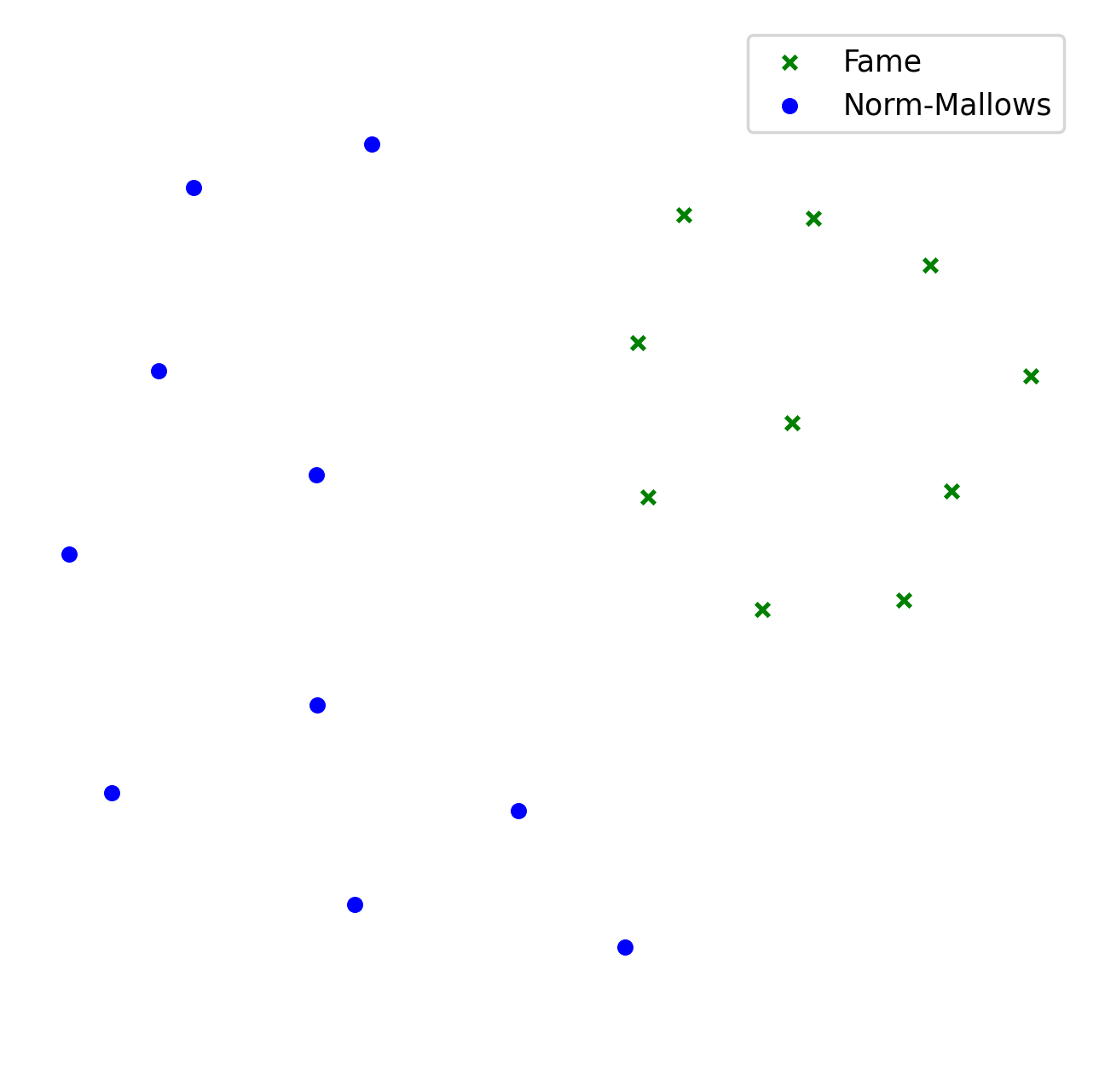
Example 2.#
experiment = mapof.prepare_online_roommates_experiment()
experiment.set_default_num_agents(20)
experiment.add_family(culture_id='fame', size=10)
experiment.add_family(culture_id='norm-mallows', size=10, normphi=0.33)
experiment.compute_distances(distance_id='l1-mutual_attraction')
experiment.embed_2d(embedding_id='kk')
experiment.print_map_2d()
As a result of the code above, you will see two separate black clouds of points (see Example 1.). In order to make the map more pleasing, we can specify the colors/markers/label of each instance or family of instances separately. We do it via color, marker, label arguments.
experiment = mapof.prepare_online_roommates_experiment()
experiment.set_default_num_agents(20)
experiment.add_family(culture_id='fame', size=10,
color='green', marker='x', label='Fame')
experiment.add_family(culture_id='norm-mallows', size=10,
normphi=0.33,
color='blue', marker='o',
label='Norm-Mallows')
experiment.compute_distances(distance_id='l1-mutual_attraction')
experiment.embed_2d(embedding_id='kk')
experiment.print_map_2d()
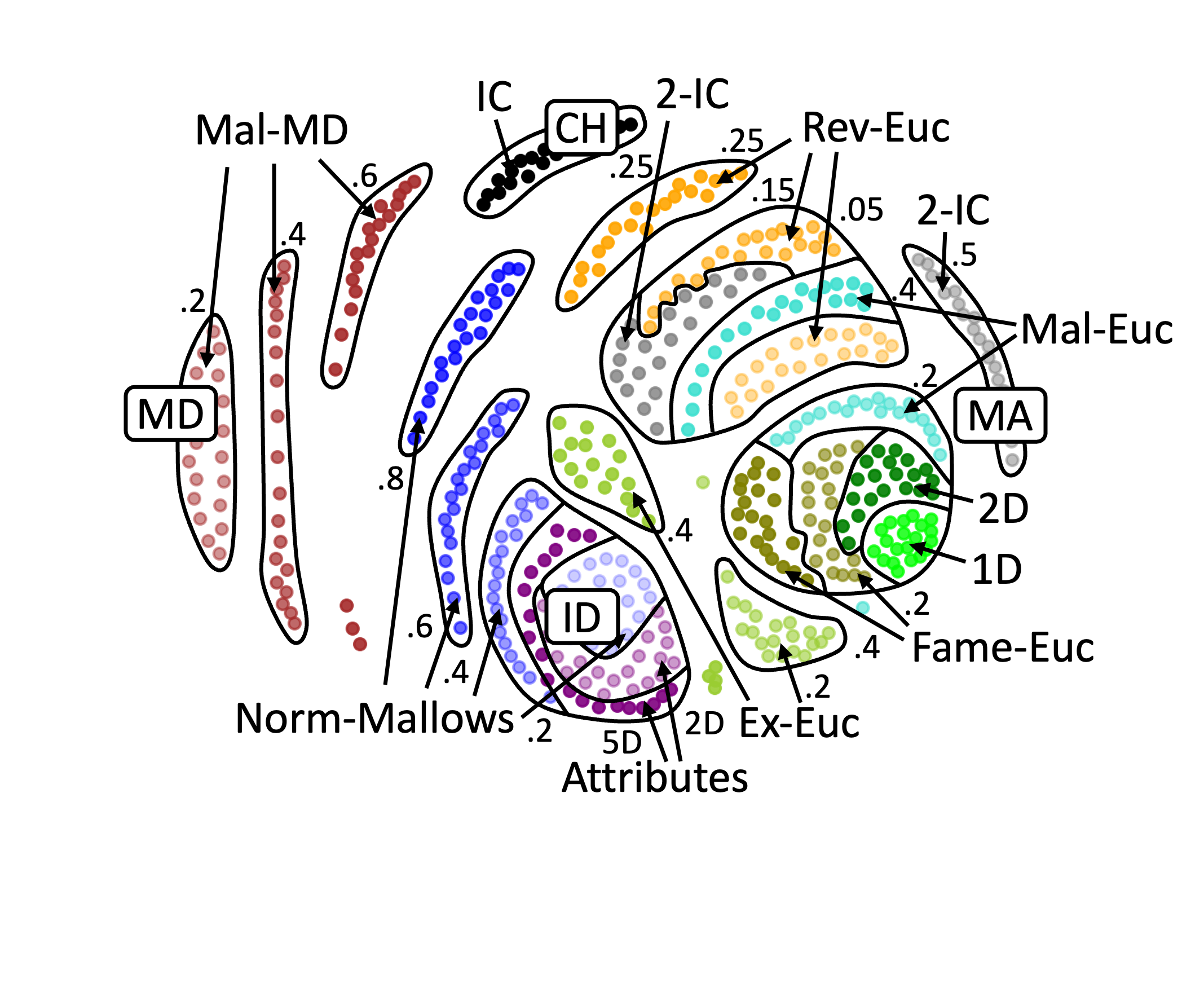
Example 3: A map for the 200 dataset of Böhmer et al. [2024].#
The picture created by the improved version is presented in Example 2.. Moreover, for illustrative purposes, in Example 3 we present the map for the 200 dataset of Böhmer et al. [2024]. Note that the labels and arrows are created in PowerPoint and are not part of the mapof software.
Coloring Map of SR Instances#
It is interesting to color the map according to certain statistics, referred to as features.
Basic
We offer several pre-implemented features. For example, if you would like to compute the highest plurality score for all instances, you can write:
experiment.compute_feature(feature_id='mutuality')
To print it, use the feature argument:
experiment.print_map_2d_colored_by_feature(feature_id='mutuality')
To access the computed values, type:
experiment.features['mutuality']
List of all the available features can be found in the List of Features.
Printing
Basic arguments for the print_map_2d function are the following:
saveas=str # save file as xyz.png
title=str # title of the image
legend=bool # (by default True) if False then hide the legend
ms=int # (by default 20) size of the marker
show=bool # (by default True) if False then hide the map
cmap # cmap (only for printing features)
For example:
experiment.print_map_2d(title='My First Map', saveas='tmp', ms=30)
Offline Experiment#
Offline experiments are similar to online experiments but offer the possibility to export/import files with instances, distances, coordinates, features, etc.
Prepare Experiment
To prepare an offline experiment, run:
experiment = mapof.prepare_offline_roommates_experiment(
experiment_id='name_of_the_experiment')
The function above will create the experiment structure as follows:
experiment_id/
├── coordinates/
├── distances/
├── instances/
├── features/
└── map.csv
Prepare Instances
To prepare instances, run:
experiment.prepare_instances()
Instances are generated according to the map.csv file. An example map.csv file is created automatically when preparing the experiment.
map.csv
The controlling map.csv file usually consists of:
size: Number of instances to be generated from a given culture
num_agents: Number of candidates
culture_id: Code of the culture
params: Dictionary with parameters of a given culture
color: Color of the point(s) on the map
alpha: Transparency of the point(s)
marker: Marker of the point(s)
ms: Marker size
label: Label that will be printed in the legend
family_id: Family ID
path: Dictionary with parameters for generating a path of instances
Imports
If some parts of your experiment are already precomputed, you can import them while preparing the experiment. Ensure they are in the proper files. If they were precomputed using mapof, no additional steps are required.
If you want to import specific elements (different from default), specify them while preparing the experiment. For transparency, it is recommended to always define them.
experiment = mapof.prepare_offline_ordinal_experiment(
experiment_id='name_of_the_experiment',
distance_id="l1-mutual_attraction'",
embedding_id="kk")
Regarding features, if they are precomputed, the program will import them while printing the map.